To simplify dendritic spine analysis, our histology and IT team joined forces and developed a macro-based image analysis for rater-independent, unbiased, fast, and efficient automated dendritic spine quantification.
Brain sections are stained by the Golgi-Cox method. For spine density analysis, Z-stack images are taken using 63x oil lens, and stacks are collapsed using Zeiss ZEN software. Spines are detected and quantified automatically by a macro running in Image Pro 10 software.
Spine density analyses are of interest for studies associated with different human diseases and their animal models:
- Spine density analysis in the SOD1G93A mouse model of amyotrophic lateral sclerosis. Fogarty et al. 2016. https://doi.org/10.1186/s40478-016-0347-y
- Dendritic spine remodeling accompanies Alzheimer’s disease pathology and genetic susceptibility in cognitively normal aging. Boros et al. 2019. https://doi.org/10.1016/j.neurobiolaging.2018.09.003
- Common defects of spine dynamics and circuit function analyzed in neurodevelopmental disorders, including BTBR T+ tf/J mice as an inbred strain model of autism spectrum disorder. Nakai et al. 2018. https://doi.org/10.3389/fnins.2018.00412
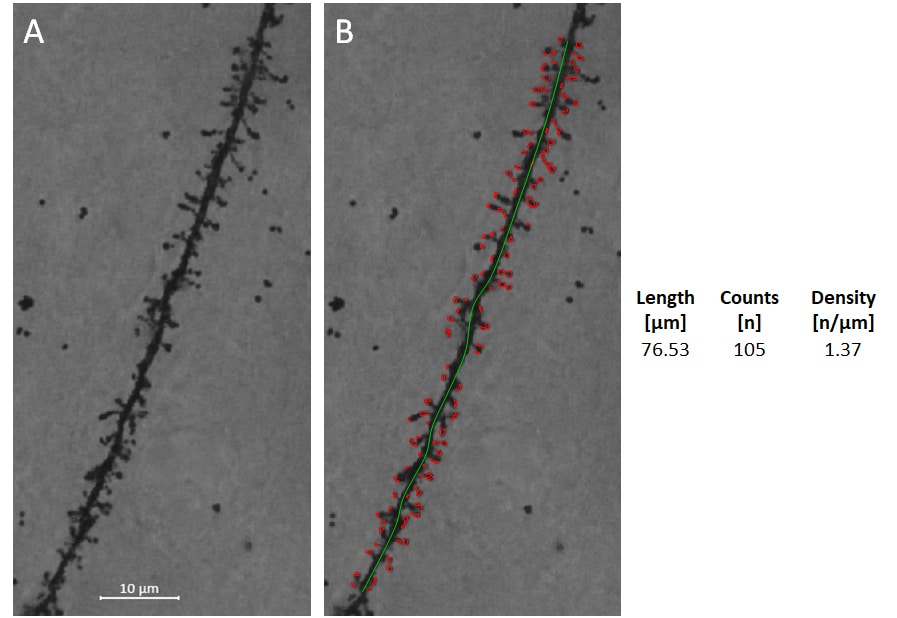
Figure: Spine recognition and spine density analysis. A: Collapsed Z-stack image of Golgi-Cox stained dendrite of frontal cortical pyramidal cell. B: Length of the dendritic segment was determined (green line), and spines were then identified and counted automatically. The data generated from this image are shown on the right.
Contact us today to get your study tissue evaluated for Golgi-Cox staining!









